Dynamics of Cell Biology in the Germline
We have begun to define in vivo protein-protein interaction networks, concentrating on proteins with important roles in the early embryo. A major focus of our work has been to characterize novel proteins required for germline stem cell maintenance in C. elegans. Germ granules, known as P granules in C. elegans, localize specifically to the germ cell lineage throughout development. In the adult germline, P granules are the sites of small RNA processing, which is needed to maintain genome integrity and memory of the germline state across generations. P granules also regulate the stability and translation of mRNAs that are needed for key developmental events.
Membraneless organelles such as P granules, stress granules, and even nucleoli have recently garnered much attention in the last few years because they form dynamic phase-separated compartments with interesting biophysical properties that are involved in many aspects of cell biology. Phase separation is thought to increase the efficiency of molecular processes by concentrating specific components while protecting them from other cellular factors (Shin and Brangwynne, Science 2017). We’ve discovered new novel components of P granules and are currently investigating their roles in developmental processes, as well as their interactions with other granule factors to regulate germ granule assembly, localization, and function (Cipriani et al., eLife 2021; Marnik et al, PLOS Genetics 2022).

Evolution of Early Development and Reproductive Models
Sexual reproduction provides survival advantages by promoting genetic diversity and limiting the accumulation of deleterious alleles within populations. Thus parthenogenesis, in which oocytes produce progeny in the absence of fertilization, is usually a dead-end reproductive strategy that leads to short-lived lineages. Nevertheless, parthenogenesis has evolved multiple times independently across different animal clades. Studying genetic mechanisms that can give rise to such changes can provide a deeper understanding of how reproductive programs can evolve.
We have sequenced the genome of Diploscapter pachys, a distant relative of C. elegans belonging to an unusually long-lived clade of parthenogenetic nematodes (Fradin et al., Current Biology 2017). D. pachys harbors only a single giant chromosome that we showed derives from the fusion of all ancestral chromosomes. We showed that D. pachys successfully reproduces asexually by skipping the reductional meiotic division, Meiosis I. This work is leading to mechanistic insights into the evolution of sexual reproduction, meiotic recombination, and establishment of the body plan in early development. We are now extending this study to closely related species to learn more about the evolutionary changes that took place in this lineage, and to further understand the molecular processes driving chromosome fusion and developmental changes that accompany this change in reproductive strategy.
Genetic Interaction Networks
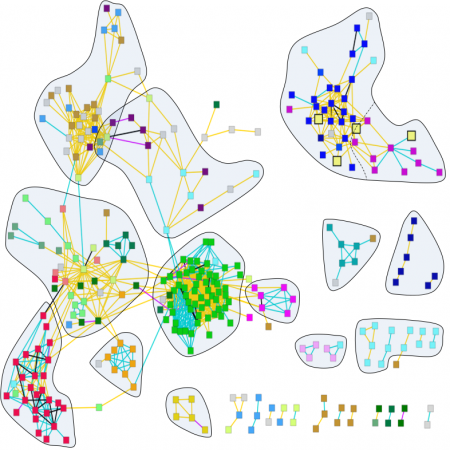
We have undertaken a major effort to systematically investigate how essential and non-essential genes in C. elegans interact with each other. We have developed a high-throughput platform to analyze interactions between all genes and 24 temperature-sensitive (ts) alleles of genes with essential roles in diverse processes in the early embryo. We have so far assembled the results from over 5 million experiments and have begun to analyze them. Current results show that certain metabolic products have previously unrecognized roles in very early development and, more generally, open a new window to study how metabolic pathways are integrated with developmental processes.
How do molecular networks evolve in time and space? Understanding the dynamics of molecular interactions and how these relate to complex traits is now a central focus in the field of systems biology that is being pursued from many angles by the research community. To address this question, we conducted over 5 million experiments testing interactions between genes with roles in early development of C. elegans, and are now analyzing the results.
Post-Transcriptional Regulation
RNA regulation is a continuing focus in the lab. In a survey of 3’UTR dynamics in the germline, we discovered a master regulator of ribosome biogenesis, the protein NCL-1 (West et al., Genome Biol 2018). NCL-1 has recently been shown to extend longevity (Tiku et al., Nat Comm 2017). We think NCL-1 likely integrates dietary status with metabolic programs, and we are working to understand what signals NCL-1 senses and how it mediates its downstream effects.
We also use 3D structure modeling in order to understand more fully the basis of RNA recognition by proteins (Gan et al, Frontiers in Virology 2022 and J Mol Biol 2021). A particular focus has been the microRNA-silencing complex (miRISC), which comprises an Argonaute protein bound to a miRNA-target RNA duplex. Using a modeling pipeline we developed, our models have revealed how specific structural and conformational features contribute to miRISC binding in ways that cannot be determined by primary and secondary structure analysis alone (Gan and Gunsalus, Methods Mol Biol 2019).