Recent News
The Caenorhabditis elegans TDRD5/7-like protein, LOTR-1, interacts with the helicase ZNFX-1 to balance epigenetic signals in the germline
In our most recent paper in PLOS Genetics, we show that LOTR-1 partitions with a specific class of subgranules called Z granules, interacting with a Z-granule helicase called ZNFX-1. Here, LOTR-1 functions with ZNFX-1 to position small RNA amplification from RNA templates, ensuring a memory of germline expression across generations. These findings may provide new insight into the function of TDRD5 and TDRD7 during human germline development.
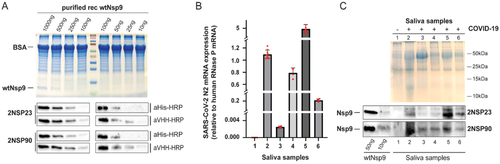
NMR‐Based Analysis of Nanobodies to SARS‐CoV‐2 Nsp9 Reveals a Possible Antiviral Strategy Against COVID‐19
New nanobodies targeting Nsp9, a nonstructural protein required for Sars‐CoV‐2 replication, are generated. These nanobodies specifically recognize Nsp9 in saliva samples from COVID‐19 patients and stabilize a tetrameric form of Nsp9, not compatible with its role in facilitating viral replication. These results potentially identify novel diagnostic and therapeutic antiviral strategies to combat Sar‐CoV‐2 and, therefore, COVID‐19. Read more in Advanced Biology.
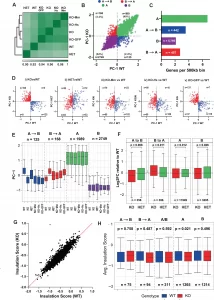
β-actin dependent chromatin remodeling mediates compartment level changes in 3D genome architecture
We demonstrate in Nature Communications that β-actin levels can induce changes in chromatin structure by affecting the complex interplay between chromatin remodelers such as BAF/BRG1 and EZH2. Our results show that changes in β-actin levels and associated chromatin remodeling activities can not only impact local chromatin accessibility but also induce reversible changes in 3D genome architecture. Our findings reveal that β-actin-dependent chromatin remodeling plays a role in shaping the chromatin landscape and influences the regulation of genes involved in development and differentiation.
Novel LOTUS-domain proteins are organizational hubs that recruit C. elegans Vasa to germ granules
In this paper, published in eLife, we describe MIP-1 and MIP-2, novel paralogous C. elegans germ granule components that interact with the intrinsically disordered MEG-3 protein.We propose that the MIP proteins serve as scaffolds and organizing centers for ribonucleoprotein networks within P granules that help recruit and balance essential RNA processing machinery to regulate key developmental transitions in the germ line.
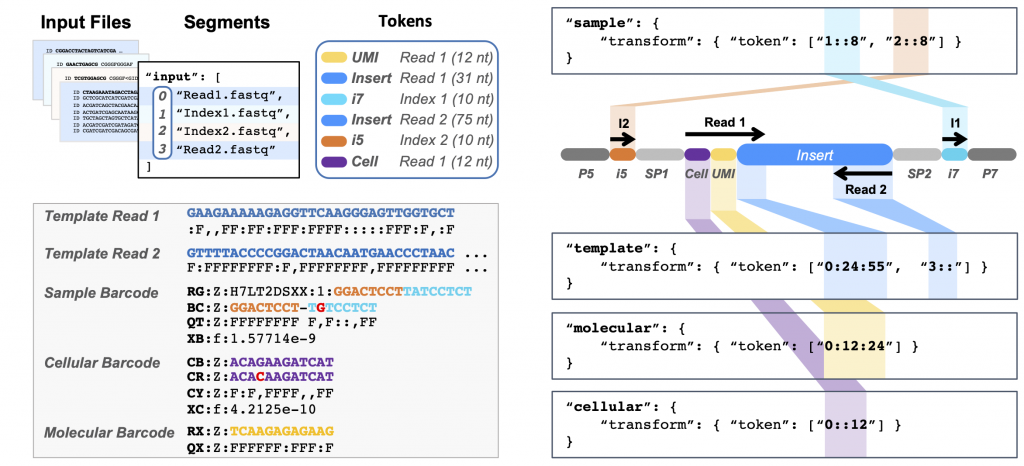
Pheniqs 2.0: accurate, high-performance Bayesian decoding and confidence estimation for combinatorial barcode indexing
We introduce a computationally efficient software that implements both probabilistic and minimum distance decoders and show that decoding barcodes using posterior probabilities is more accurate than available methods. Pheniqs allows fine-tuning of decoding sensitivity using intuitive confidence thresholds and is extensible with alternative decoders and new error models. Read more about Pheniqs in our BMC Bioinformatics paper.
Computer-Based Modeling Can Predict Mutation “Hotspots” and Antibody Escapers in the SARS-CoV-2 Spike Protein
Our study, published in the Journal of Molecular Biology, identifies the structural basis of spike protein mutations with stronger binding and antibody resistance, which may explain transmissibility of new COVID-19 variants.